Because human immunodeficiency virus 1 (HIV-1) has an RNA genome, it is no surprise that protein-RNA interactions play key roles both in the replication cycle of HIV-1 and in the host defense against HIV-1 infection.
Research in our laboratory focuses on understanding the molecular details of how HIV-1 replication is regulated by these host and viral RNA-binding proteins, the identification of the RNA targets of these proteins, and the discovery of novel RNA-binding proteins that enhance or block HIV-1 replication.
To this end, we employ conventional and next-generation sequencing-based approaches. Currently, three main areas of focus are:
Novel role for HIV-1 integrase particle maturation
We have recently revealed an unexpected novel role for the HIV-1 integrase (IN) protein in particle maturation. In particular, we have shown that IN binds to the viral RNA genome during particle maturation and ensures that the genome is packaged in the mature capsid core. If IN-RNA interactions are blocked either through mutations within IN or by a novel class of compounds that are in clinical trials, viral RNA is localized outside the capsid lattice and the viral cores are aberrantly formed. We are currently investigating the morphological defects in these particles and what makes them non-infectious in target cells. We are also exploring how IN-RNA interactions regulate IN multimerization, which appears to be key for both particle maturation and the catalytic activity of IN.

Roles of RNA-binding proteins in HIV-1 replication
We study how several viral and cellular RNA-binding proteins help and restrict HIV-1 replication. One area of research is to understand how two copies of the viral RNA genome are selectively packaged by the Gag, HIV-1 major structural protein in virus particles. Based on our recent findings, we are in particular interested in determining whether the unusual genomic content (~40% A-rich) of HIV-1, determines the selective packaging of the viral genome. A second area of interest is to reveal novel regulatory mechanisms of HIV-1 alternative RNA splicing. A third area of interest is to determine how host cellular proteins, which are typically induced by host innate immune responses, limit HIV-1 replication. Our work in this area involves the discovery of novel interferon-indued RNA-binding proteins that limit HIV-1 replication. To address these questions, we employ cutting-edge next-generation sequencing-based approaches.

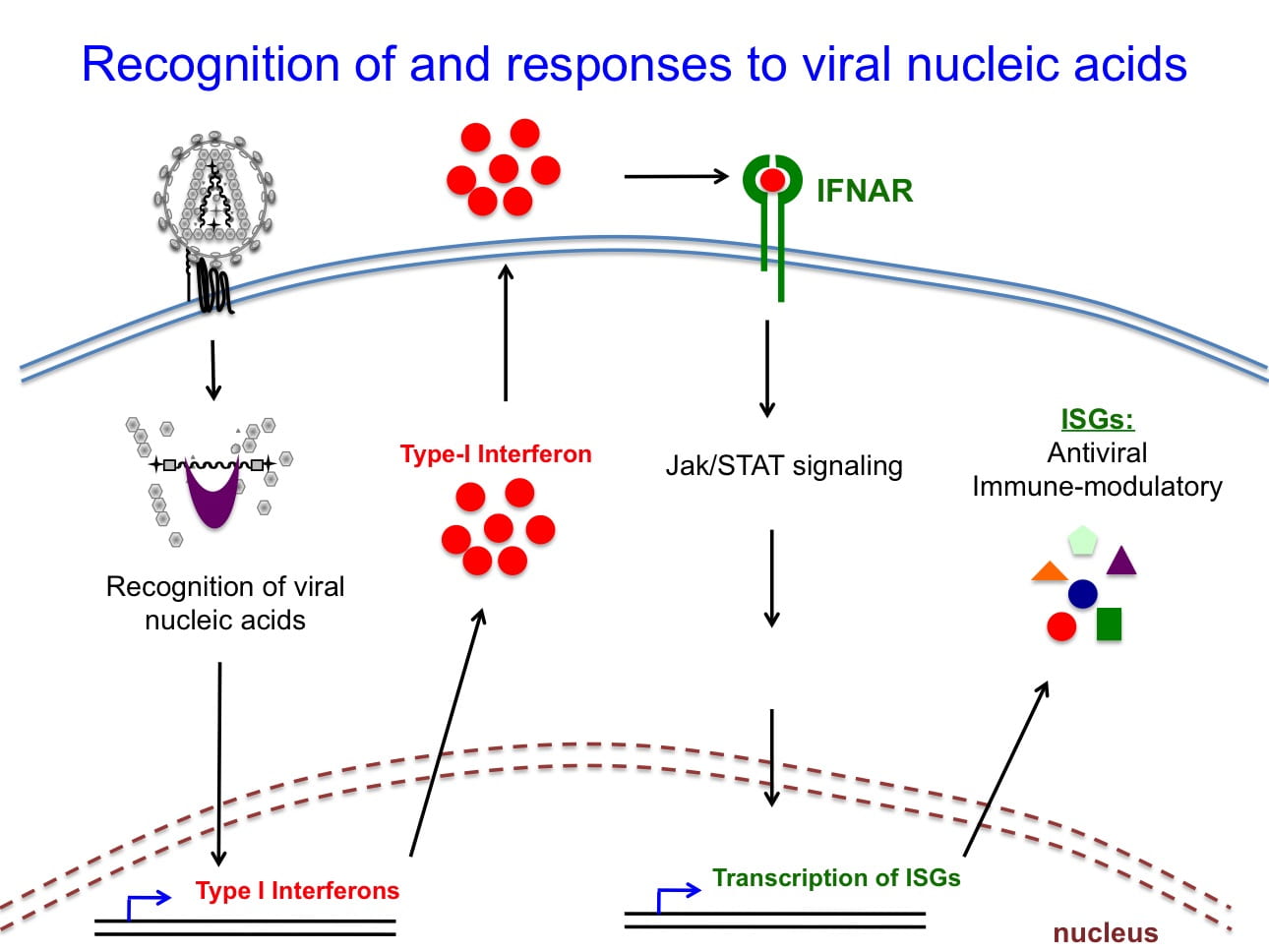
Recognition of viral nucleic acids in cells
The earliest phase of the innate immune responses against viral pathogens is mediated by a set of proteins termed pattern recognition receptors. These molecules recognize distinct elements within viral RNAs or DNAs, and activate a signaling cascade culminating in the synthesis of type-1 interferons as well as proinflammatory cytokines and chemokines that recruit and activate innate immune cells. However, there is paucity of data on whether the proposed viral sequence and structural features that constitute the recognition elements exist in infected cells. By employing CLIP-seq and related approaches, we aim to determine the sequence and structural features of HIV-1 viral nucleic acids bound by host sensors in infected cells. We are also interested in identifying novel downstream events that limit HIV-1 replication.